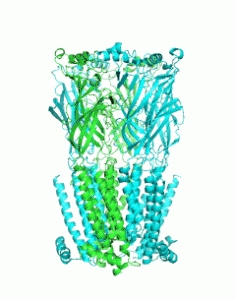
Through analysis of known protein structures, it is possible to gain insight into the rules that dictate how proteins fold. However, the number of experimentally determined protein structures is large and growing rapidly, which makes even the categorisation of protein structure difficult to perform. Computational tools can ameliorate this process, through automated categorisation and analysis.
A team from the University of Bristol, led by Prof Dek Woolfson, have recently published an article on AtlasCC. This computational resource automatically analyses the PDB to find an important protein substructure called the α-helical coiled coil and uses graph theory enumerate all the possible and observed structures this fold can adopt. These data are made accessible using a user-friendly, interactive web application that enables users to browse the structures. The application also identifies regions of coiled-coil structure space that has not been explored by nature, indicating possible opportunities for de novo design.