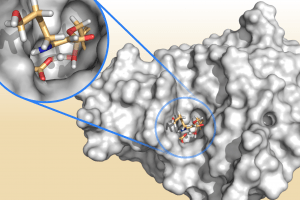
Growing resistance to antibiotics is a major threat to human health and there is an urgent need to develop new antibiotics or find new ways to keep existing antibiotics effective. β-lactamases are a class of enzymes that have evolved to break down penicillin-type antibiotics, leading to resistance. One method to avoid this is to administer a β-lactamase inhibitor as a resistance blocker. Clavulanic acid is one such example and is often combined with amoxicillin to form the drug commonly known as co-amoxiclav. In a recent paper published in Biochemistry, van der Kamp and co-workers have used combined quantum mechanics / molecular mechanics (QM/MM) methods to investigate how clavulanic acid can be broken down by different β-lactamases. TEM-1 and KPC-2 are clinically important enzymes that exhibit different susceptibilities to inhibition by clavulanic acid, with KPC-2 effectively resistant to inhibition by clavulanic acid.
QM/MM simulations were carried in AMBER with the semiempirical SCC-DFTB method used for the QM region and the MM region described by AMBER’s ff14SB force field. Complete two-dimensional free energy surfaces were then obtained by QM/MM umbrella sampling (US) simulations along two reaction coordinates: one to describe the proton transfer and one to describe the nucleophilic attack involved in deacylation step of the reaction. The weighted-histogram analysis method (WHAM) was used to generate the free energy surfaces and the minimum free energy path (MEP) on the surfaces was determined using the minimum energy pathway analysis for energy landscapes (MEPSA) software. The results show that the QM/MM screening protocol is reliable in discriminating the inhibitory activity of different covalent complexes in class A β-lactamases and can differentiate enzymes for which clavulanic acid is an effective inhibitor (TEM-1) from those for which it is not (KPC-2). Such screening techniques will prove to be useful in the development of new strategies to overcome antibiotic resistance.
Press release:
http://www.bristol.ac.uk/news/2018/june/antibiotic-arms-race.html
Full paper:
‘Multiscale simulations of clavulanate inhibition identify the reactive complex in class A β-lactamases and predict efficiency of inhibition’ by Rubén A. Fritz, Jans H. Alzate-Morales, James Spencer, Adrian J. Mulholland and Marc W. van der Kamp in Biochemistry.