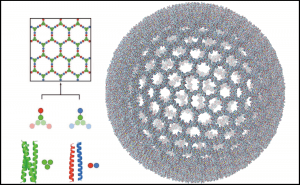
Our manuscript entitled, “The dynamical interplay between a megadalton peptide nanocage and solutes probed by microsecond atomistic MD; implications for design” has been accepted for publication in Physical Chemistry Chemical Physics (PCCP), (2018), DOI: 10.1039/c8cp06282j. Deborah K. Shoemark, Amaurys Avila Ibarra, James F. Ross, Joseph L. Beesley, Harriet E.V. Bray, Majid Mosayebi, Noah Linden, Tanniemola B. Liverpool, Simon N. McIntosh-Smith, Derek N. Woolfson and Richard B. Sessions.
Here we present the lessons learnt from performing atomistic simulations of 0.6 – 1 microseconds of three ~42 million atom peptide nanocage (SAGE) systems, with and without protein and small molecule solutes. Using GROMACS and the UK supercomputer Archer, data were collected over 2 years. Detailed analysis reveals the structural integrity/helical stability of the SAGEs themselves, how SAGEs interact with the proteins and small molecule species in the systems, whether contact with SAGE influences the native fold of solute proteins and how frequently solutes or ions pass through pores in the SAGEs. Knowing how and where to elaborate and/or modify the SAGE building blocks is important to inform the design process for this synthetic biological approach to diverse applications. As a vaccine delivery platform, SAGEs that are modular and reproducible, could be tailored to respond to emerging infectious threats more rapidly than conventional methods. For targeted drug delivery, adding peptides that bind SAGEs packed with drug, to specific cell types could reduce dosage demand and side-effects. As nanoreactors for complex chemistry, encapsulating enzyme pathways may allow for more efficient and environmentally friendly catalysis.